Mitotic counting via deep learning and manual assessment is not prognostic in TNBC.
The prognostic value of mitotic count for invasive breast cancer is firmly established. However, to date, no studies have been reported assessing the prognostic value of mitotic counts in triple negative breast cancer (TNBC). TNBC tumours display wide ranges of mitotic counts with most tumours showing counts that largely exceed the minimum number required for grade 3 of the modified Bloom and Richardson grading system. Considering this wide range of mitotic figures present in TNBC, it may be questioned whether the cut-off values of the modified Bloom and Richardson grading system are applicable to TNBC or whether better suited TNBC-specific cut-off values are available. The application of deep learning would allow for a comprehensive analysis of absolute mitotic counts, even in the presence of very high densities of such cells. Therefore, Computational patholoy group-member Maschenka Balkenhol and colleagues assessed the prognostic value of absolute mitotic counts for TNBC in a retrospective study (n=298), using both deep learning and manual procedures.
The absolute manual mitotic count was assessed by averaging counts from three independent observers. Deep learning was performed using a previously described convolutional neural network on digitized H&E slides. Multivariable Cox regression models for relapse-free survival and overall survival served as baseline models. These were expanded with dichotomized mitotic counts, attempting every possible cut-off value, and evaluated by means of the c-statistic. These results are depicted below.
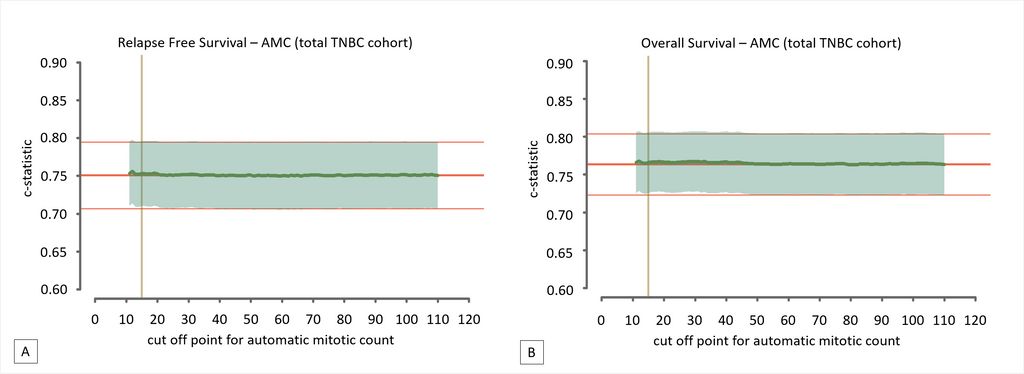
The authors found that per 2 mm2 averaged manual mitotic counts ranged from 1 to 187 (mean 37.6, SD 23.4), whereas automatic counts ranged from 1 to 269 (mean 57.6; SD 42.2). None of the cut-off values improved the models’ baseline c-statistic, for both manual and automatic assessments. This led to the conclusion that mitotic count does not serve as a prognostic factor for TNBC.
Read more about the work of Maschenka Balkenhol et al in their open access paper, that appeared online this week in Cellular Oncology.
← Back to overview