Neural Image Compression for Gigapixel Histopathology Image Analysis
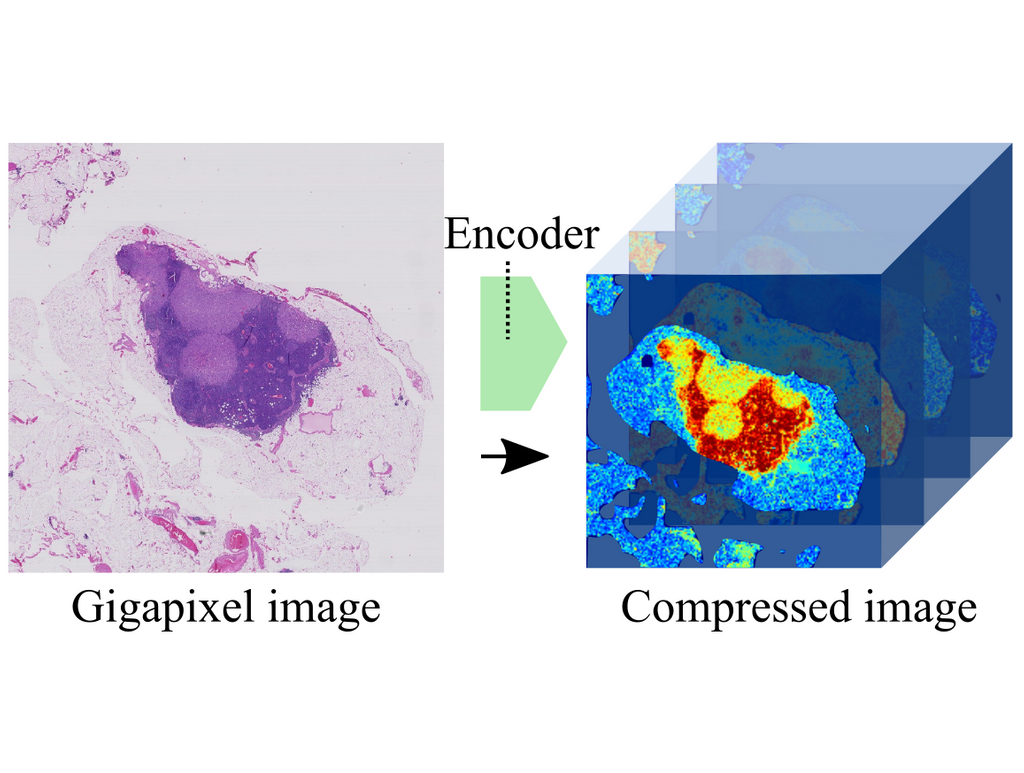
Digitized H&E slides, also known as whole-slide images (WSI), are gigantic images composed of more than 1 billion pixels. Each WSI contains a tissue sample corresponding to a single patient, describing tissue morphology to the cell level with remarkable microscopic resolution, including thousands of cells. Until now, Computational Pathology algorithms were limited to applications where fine-grained annotations were available, such as semantic segmentation or object detection.
In the work, published online in IEEE Transactions on Pattern Analysis and Machine Intelligence yesterday, the authors present a novel algorithm that can work with entire gigapixel WSIs directly, avoiding the need for fine-grained annotations. The method, name neural image compression (NIC), opens the door to training neural networks using slide-level annotations obtained automatically, e.g., targeting molecular biomarkers or patient outcome. This approach allows the neural network to discover previously unknown visual features that are relevant to predict the target at hand. NIC works in two steps. First, gigapixel images are compressed using a neural network trained in an unsupervised fashion, summarizing the image in a very efficient manner; reducing its size drastically while retaining most semantic information. Second, a convolutional neural network (CNN) is trained on these compressed image representations to predict image-level labels, avoiding the need for fine-grained manual annotations.
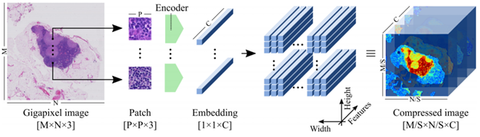
David Tellez et al compared several encoding strategies, namely reconstruction error minimization, contrastive training and adversarial feature learning, and evaluated NIC on a synthetic task and two public histopathology datasets. They found that NIC can exploit visual cues associated with image-level labels successfully, integrating both global and local visual information. Furthermore, they could visualize the regions of the WSIs where the classifier attended to, and confirmed that they overlapped with annotations from human experts.
← Back to overview