Meyke Hermsen, Thomas de Bel, Marjolijn den Boer, Eric Steenbergen, Jesper Kers, Sandrine Florquin, Joris Roelofs, Mark Stegall, Mariam Alexander, Byron Smith, Bart Smeets, Luuk Hilbrands, Jeroen van der Laak
Abstract
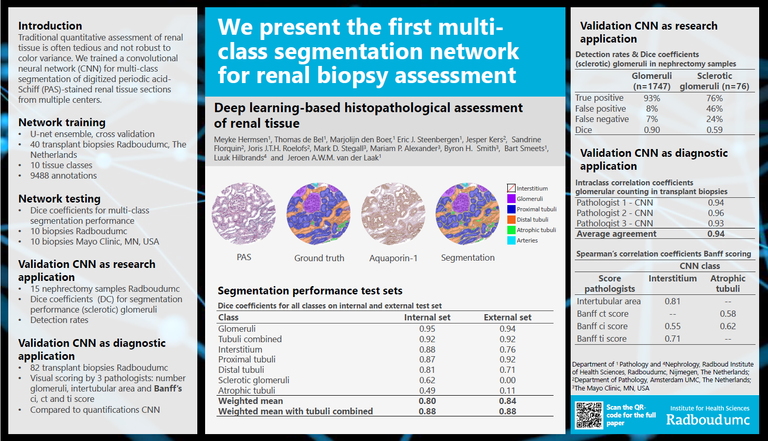
Traditional quantitative assessment of renal tissue is often tedious and not robust to color variance. We trained a convolutional neural network (CNN) for multi-class segmentation of digitized periodic acid-Schiff (PAS)-stained renal tissue sections from multiple centers.
The CNN was trained using multi-class annotations from 40 whole-slide images (WSIs) of PAS-stained renal transplant biopsies. Multi-class segmentation performance was assessed by calculating Dice coefficients (DCs) for 10 tissue classes on 10 transplant biopsies from Radboudumc and on 10 transplant biopsies from the Mayo Clinic for validation. The weighted mean DCs of all classes were 0.80 and 0.84 in 10 kidney transplant biopsies from Radboudumc and the Mayo Clinic, respectively. The best segmented class was ‘glomeruli’ in both data sets (DC 0.95 and DC 0.94), followed by ‘tubuli combined’ and ‘interstitium’. Additionally, we fully segmented 15 tumor nephrectomy samples and assessed the CNN’s glomerular detection rates. The CNN detected 92.7% of all glomeruli in nephrectomy samples, with 10.4% false positives. Lastly, CNN-based measures were compared with visually scored histological (Banff) components in 82 kidney transplant biopsies. The mean intraclass correlation coefficient for glomerular counting performed by pathologists versus the CNN was 0.94. Significant correlations were observed between relevant components of the visual Banff scoring system and CNN-based measures.
This study presents the first CNN for multi-class segmentation of PAS-stained nephrectomy samples and transplant biopsies. Our CNN can be of aid for quantitative studies concerning renal histopathology across centers and provides opportunities for deep learning applications in routine diagnostics.