Most men die with, not because of prostate cancer. This high incidence-to-mortality ratio sounds like a positive trait, but comes with its own share of problems: high risk of overdiagnosis and overtreatment with significant patient morbidity. To combat overtreatment, several models have been developed to assign patients to risk categories with differing treatment regimes. Although these models show good correlation with patient outcome on the group level, their benefit for the individual patient remains limited.
Several groups have shown that quantifying the tumour and its micro-environment at the cellular level can result in biomarkers with strong prognostic potential, for example tumour/stroma ratio, the presence and composition of immune infiltrates or the amount of proliferating (Ki67-positive) cells. However, these findings have not translated to clinical practice due to the cumbersome and subjective manual extraction of these biomarkers from tissue slides.
Recent years have seen the more widespread introduction of whole-slide imaging systems, which for the first time allow computerized processing of tissue slides. Automated extraction of aforementioned quantitative biomarkers through image analysis can achieve the required accuracy and robustness to impact clinical practice. In tandem with these developments, computer vision has seen a machine learning revolution over the past decade due to the advent of deep learning.
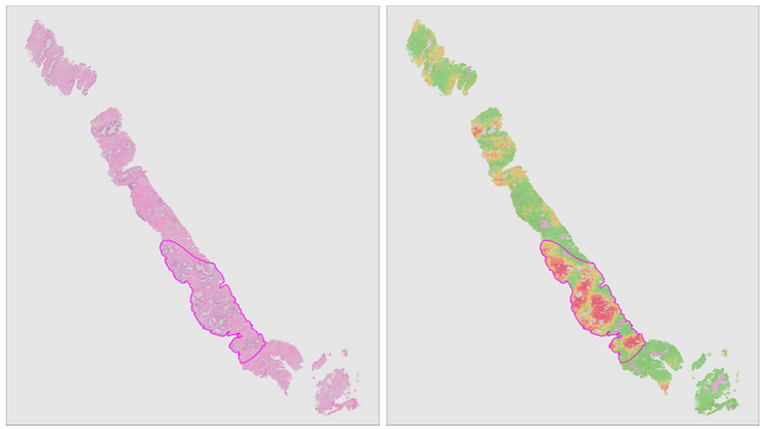
In this project, we will combine deep learning and digitized whole-slide imaging of prostate cancer for reproducible extraction of quantitative biomarkers. Furthermore, due to the ability of deep learning systems to learn relevant features without human intervention, we expect to identify novel biomarkers which allow us to further improve the current risk models.
The aim of this project is to prevent unnecessary surgery and adjuvant therapy for individual patients by improving currently established risk models. Risk models will be enhanced through the addition of pre- and post-operative quantitative biomarkers obtained via image analysis and deep learning. We will focus both on the accurate and objective quantification of biomarkers already identified in literature and the discovery of novel biomarkers.





