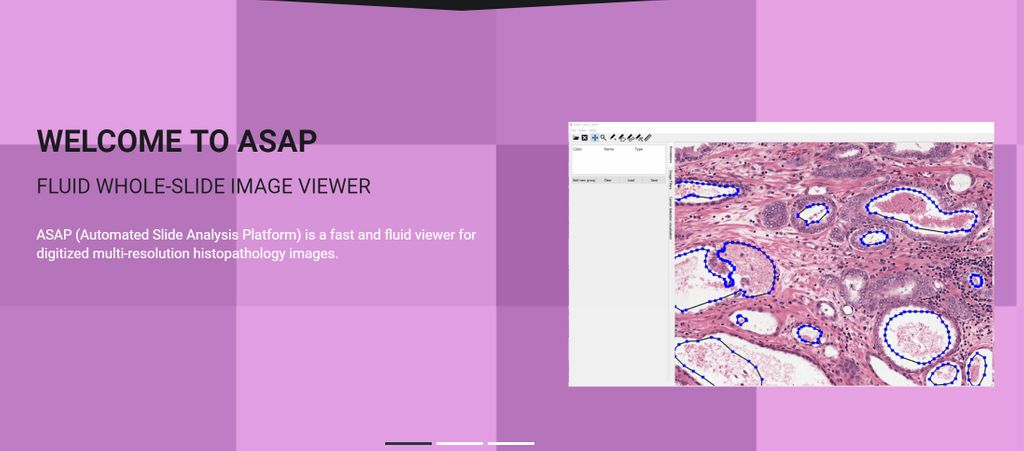
This project is aimed to develop open-source software for handling the large images resulting from scanning whole pathology slides (whole slide images; WSI) at microscopic resolution. The software enables researchers and pathologists to view WSI, similar to the visual inspection of slides under a microscope. To this end, the software employs Google-earth type of functionality, enabling seamless panning and zooming of the images. In addition, functionality for annotating images was developed and a plug-in interface was generated to enable use of algorithms from 'third parties'. The resulting 'automated slide analysis platform' (ASAP) open source software is capable of dealing with WSI from almost all scanners currently in use.
In addition, within this project we studied the possibility to automatically detect metastases in WSI of sentinel lymph nodes of breast cancer patients. Next to use of traditional machine learning algorithms, we also studied the use of (at that time still relatively unknown) deep learning (DL) technology. This resulted in one of the first publications describing the large potential of DL for analyzing entire WSI as an aid to pathology diagnostics. Subsequently, we extended this study and organized the CAMELYON16 'grand challenge'. To date, this is the most influential grand challenge organized in pathology, with a publication in JAMA as well as extensive media coverage.

